Conformational change
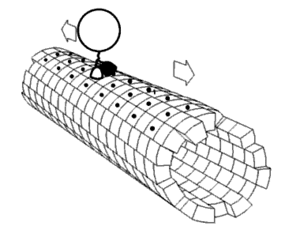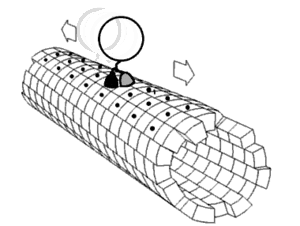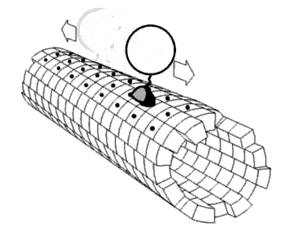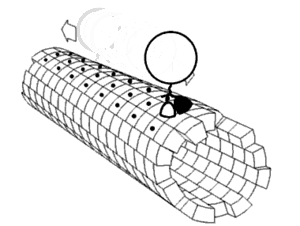
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors.
A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a conformational change. Factors that may induce such changes include temperature, pH, voltage, light in chromophores, concentration of ions, phosphorylation, or the binding of a ligand. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling[1] and enzyme catalysis.[2]
Laboratory analysis[edit]
Many biophysical techniques such as crystallography, NMR, electron paramagnetic resonance (EPR) using spin label techniques, circular dichroism (CD), hydrogen exchange, and FRET can be used to study macromolecular conformational change. Dual-polarization interferometry is a benchtop technique capable of providing information about conformational changes in biomolecules.[3]
A specific nonlinear optical technique called second-harmonic generation (SHG) has been recently applied to the study of conformational change in proteins.[4] In this method, a second-harmonic-active probe is placed at a site that undergoes motion in the protein by mutagenesis or non-site-specific attachment, and the protein is adsorbed or specifically immobilized to a surface. A change in protein conformation produces a change in the net orientation of the dye relative to the surface plane and therefore the intensity of the second harmonic beam. In a protein sample with a well-defined orientation, the tilt angle of the probe can be quantitatively determined, in real space and real time. Second-harmonic-active unnatural amino acids can also be used as probes.[citation needed]
Another method applies electro-switchable biosurfaces where proteins are placed on top of short DNA molecules which are then dragged through a buffer solution by application of alternating electrical potentials. By measuring their speed which ultimately depends on their hydrodynamic friction, conformational changes can be visualized.[citation needed]
"Nanoantennas" made out of DNA – a novel type of nano-scale optical antenna – can be attached to proteins and produce a signal via fluorescence for their distinct conformational changes.[5][6]
Computational analysis[edit]
X-ray crystallography can provide information about changes in conformation at the atomic level, but the expense and difficulty of such experiments make computational methods an attractive alternative.[7] Normal mode analysis with elastic network models, such as the Gaussian network model, can be used to probe molecular dynamics trajectories as well as known structures.[8][9] ProDy is a popular tool for such analysis.[10]
Examples[edit]
Conformational changes are important for:
- ABC transporters [11]
- catalysis[12]
- cellular locomotion and motor proteins[13]
- formation of protein complexes[14]
- ion channels[15]
- mechanoreceptors and mechanotransduction[16]
- regulatory activity [17]
- transport of metabolites across cell membranes [18][19]
See also[edit]
- Database of protein conformational diversity
- Protein dynamics
- The Database of Macromolecular Motions (molmovdb)
References[edit]
- ^ Bu Z, Callaway DJ (2011). "Proteins move! Protein dynamics and long-range allostery in cell signaling". Protein Structure and Diseases. Vol. 83. pp. 163–221. doi:10.1016/B978-0-12-381262-9.00005-7. ISBN 9780123812629. PMID 21570668.
{{cite book}}:|journal=ignored (help) - ^ Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T (December 2009). "Hidden alternative structures of proline isomerase essential for catalysis". Nature. 462 (7273): 669–73. Bibcode:2009Natur.462..669F. doi:10.1038/nature08615. PMC 2805857. PMID 19956261.
- ^ Freeman NJ, Peel LL, Swann MJ, Cross GH, Reeves A, Brand S, Lu JR (2004-06-19). "Real time, high resolution studies of protein adsorption and structure at the solid–liquid interface using dual polarization interferometry". Journal of Physics: Condensed Matter. 16 (26): S2493–S2496. Bibcode:2004JPCM...16S2493F. doi:10.1088/0953-8984/16/26/023. ISSN 0953-8984. S2CID 250737643.
- ^ Salafsky JS, Cohen B (November 2008). "A second-harmonic-active unnatural amino acid as a structural probe of biomolecules on surfaces". The Journal of Physical Chemistry B. 112 (47): 15103–7. doi:10.1021/jp803703m. PMID 18928314.
- ^ "Chemists use DNA to build the world's tiniest antenna". University of Montreal. Retrieved 19 January 2022.
- ^ Harroun, Scott G.; Lauzon, Dominic; Ebert, Maximilian C. C. J. C.; Desrosiers, Arnaud; Wang, Xiaomeng; Vallée-Bélisle, Alexis (January 2022). "Monitoring protein conformational changes using fluorescent nanoantennas". Nature Methods. 19 (1): 71–80. doi:10.1038/s41592-021-01355-5. ISSN 1548-7105. PMID 34969985. S2CID 245593311.
- ^ Kim Y, Bigelow L, Borovilos M, Dementieva I, Duggan E, Eschenfeldt W, et al. (2008-01-01). "Chapter 3. High-throughput protein purification for x-ray crystallography and NMR". Advances in Protein Chemistry and Structural Biology. 75: 85–105. doi:10.1016/S0065-3233(07)75003-9. PMC 3366499. PMID 20731990.
- ^ Tang QY, Kaneko K (February 2020). "Long-range correlation in protein dynamics: Confirmation by structural data and normal mode analysis". PLOS Computational Biology. 16 (2): e1007670. Bibcode:2020PLSCB..16E7670T. doi:10.1371/journal.pcbi.1007670. PMC 7043781. PMID 32053592.
- ^ Zheng W, Doniach S (November 2003). "A comparative study of motor-protein motions by using a simple elastic-network model". Proceedings of the National Academy of Sciences of the United States of America. 100 (23): 13253–8. Bibcode:2003PNAS..10013253Z. doi:10.1073/pnas.2235686100. PMC 263771. PMID 14585932.
- ^ Bakan A, Meireles LM, Bahar I (June 2011). "ProDy: protein dynamics inferred from theory and experiments". Bioinformatics. 27 (11): 1575–7. doi:10.1093/bioinformatics/btr168. PMC 3102222. PMID 21471012.
- ^ Ponte-Sucre A, ed. (2009). ABC Transporters in Microorganisms. Caister Academic. ISBN 978-1-904455-49-3.
- ^ Kamerlin SC, Warshel A (May 2010). "At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?". Proteins. 78 (6): 1339–75. doi:10.1002/prot.22654. PMC 2841229. PMID 20099310.
- ^ Howard J (2001). Mechanics of motor proteins and the cytoskeleton (1st ed.). Sunderland,MA: Sinauer Associates. ISBN 9780878933334.
- ^ Callaway DJ, Matsui T, Weiss T, Stingaciu LR, Stanley CB, Heller WT, Bu Z (April 2017). "Controllable Activation of Nanoscale Dynamics in a Disordered Protein Alters Binding Kinetics". Journal of Molecular Biology. 429 (7): 987–998. doi:10.1016/j.jmb.2017.03.003. PMC 5399307. PMID 28285124.
- ^ Hille B (2001) [1984]. Ion Channels of Excitable Membranes (3rd ed.). Sunderland, Mass: Sinauer Associates, Inc. p. 5. ISBN 978-0-87893-321-1.
- ^ Nicholl ID, Matsui T, Weiss TM, Stanley CB, Heller WT, Martel A, et al. (August 2018). "α-Catenin Structure and Nanoscale Dynamics in Solution and in Complex with F-Actin". Biophysical Journal. 115 (4): 642–654. Bibcode:2018BpJ...115..642N. doi:10.1016/j.bpj.2018.07.005. hdl:2436/621755. PMC 6104293. PMID 30037495.
- ^ Donald V (2011). Biochemistry. Voet, Judith G. (4th ed.). Hoboken, NJ: John Wiley & Sons. ISBN 9780470570951. OCLC 690489261.
- ^ Kimball's Biology pages Archived 2009-01-25 at the Wayback Machine, Cell Membranes
- ^ Singleton P (1999). Bacteria in Biology, Biotechnology and Medicine (5th ed.). New York: Wiley. ISBN 978-0-471-98880-9.
